Classification of Various Marijuana Varieties by Raman Microscopy and Chemometrics
Abstract
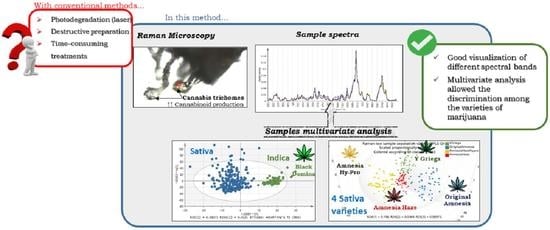
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Samples
2.2. Quantum Chemical Simulated Raman Spectra
2.3. Raman Analysis and Spectra Acquisition
2.4. Data Pre-Processing and Analysis
3. Results and Discussion
3.1. Marijuana Spectral Characteristics
3.2. Marijuana Classification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andre, C.M.; Hausman, J.-F.; Guerriero, G. Cannabis sativa: The Plant of the Thousand and One Molecules. Front. Plant Sci. 2016, 7, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsohly, M.A.; Slade, D. Chemical constituents of marijuana: The complex mixture of natural cannabinoids. Life Sci. 2005, 78, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Lucas, C.J.; Galettis, P.; Schneider, J. The pharmacokinetics and the pharmacodynamics of cannabinoids. Br. J Clin. Pharm. 2018, 84, 2477–2482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ujváry, I.; Hanuš, L. Human Metabolites of Cannabidiol: A Review on Their Formation, Biological Activity, and Relevance in Therapy. Cannabis Cannabinoid Res. 2016, 1, 90–101. [Google Scholar] [CrossRef] [Green Version]
- Substance Abuse and Mental Health Services Administration (SAMHSA). States Categorized into Five Groups from Lowest to Highest Estimate, by Age Group: Percentages, Annual Averages Based on 2019 and 2020 NSDUHs. Jama 2021, 322, 1996–2016. [Google Scholar]
- European Monitoring Centre for Drugs and Drug Addiction. European Drug Report. Trends and Developments; Publications Office of the European Union: Luxembourg, 2020. [Google Scholar]
- Kanu, A.B. Recent developments in sample preparation techniques combined with high-performance liquid chromatography: A critical review. J. Chromatogr. A 2021, 1654, 462444. [Google Scholar] [CrossRef]
- Deidda, R.; Dispas, A.; De Bleye, C.; Hubert, P.; Ziemons, É. Critical review on recent trends in cannabinoid determination on cannabis herbal samples: From chromatographic to vibrational spectroscopic techniques. Anal. Chim. Acta 2021, 33, 9184. [Google Scholar] [CrossRef]
- Sanchez, L.; Baltensperger, D.; Kurouski, D. Raman-Based Differentiation of Hemp, Cannabidiol-Rich Hemp, and Cannabis. Anal. Chem. 2020, 92, 7733–7737. [Google Scholar] [CrossRef]
- Sanchez, L.; Filter, C.; Baltensperger, D.; Kurouski, D. Confirmatory non-invasive and non-destructive differentiation between hemp and cannabis using a hand-held Raman spectrometer. RSC Adv. 2020, 10, 3212–3216. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Rev. C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Zapata, F.; López-Fernández, A.; Ortega-Ojeda, F.; Montalvo, G.; García-Ruiz, C. A practical beginner’s guide to Raman microscopy. Appl. Spectrosc. Rev. 2021, 56, 439–462. [Google Scholar] [CrossRef]
- Happyana, N.; Agnolet, S.; Muntendam, R.; Van Dam, A.; Schneider, B.; Kayser, O. Analysis of cannabinoids in laser-microdissected trichomes of medicinal Cannabis sativa using LCMS and cryogenic NMR. Phytochemistry 2013, 87, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Farag, K. Handbook of Cannabis and Related Pathologies; Elsevier, Science & Technology Books: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Turner, J.C.; Hemphill, J.K.; Mahlberg, P.G. Quantitative Determination of Cannabinoids in Individual Glandular Trichomes of Cannabis sativa L. (Cannabaceae). Am. J. Bot. 1978, 65, 1103–1106. [Google Scholar] [CrossRef]
- Ebersbach, P.; Stehle, F.; Kayser, O.; Freier, E. Chemical fingerprinting of single glandular trichomes of Cannabis sativa by Coherent anti-Stokes Raman scattering (CARS) microscopy. BMC Plant Biol. 2018, 18, 275. [Google Scholar] [CrossRef]
- Dou, T.; Sanchez, L.; Irigoyen, S.; Goff, N.; Niraula, P.; Mandadi, K.; Kurouski, D. Biochemical Origin of Raman-Based Diagnostics of Huanglongbing in Grapefruit Trees. Front. Plant Sci. 2021, 12, 41. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R; RStudio: Boston, MA, USA, 2020. [Google Scholar]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; de Jong, S.; Smilde, A.K. Direct orthogonal signal correction. Chemom. Intell. Lab. Syst. 2001, 56, 13–25. [Google Scholar] [CrossRef]
- Kvalheim, O.M.; Karstang, T.V. Interpretation of latent-variable regression models. Chemom. Intell. Lab. Syst. 1989, 7, 39–51. [Google Scholar] [CrossRef]
- Cloarec, O.; Dumas, M.E.; Craig, A.; Barton, R.H.; Trygg, J.; Hudson, J.; Blancher, C.; Gauguier, D.; Lindon, J.C.; Holmes, E.; et al. Statistical total correlation spectroscopy: An exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Anal. Chem. 2005, 77, 1282–1289. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. PCA as a practical indicator of OPLS-DA model reliability. Curr. Metab. 2016, 4, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Bylesjö, M.; Rantalainen, M.; Cloarec, O.; Nicholson, J.K.; Holmes, E.; Trygg, J. OPLS discriminant analysis: Combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 2006, 20, 341–351. [Google Scholar] [CrossRef]
- Peris-Díaz, M.D.; Krężel, A. A guide to good practice in chemometric methods for vibrational spectroscopy, electrochemistry, and hyphenated mass spectrometry. TrAC Trends Anal. Chem. 2021, 135, 116157. [Google Scholar] [CrossRef]
- Pourseyed Lazarjani, M.; Torres, S.; Hooker, T.; Fowlie, C.; Young, O.; Seyfoddin, A. Methods for quantification of cannabinoids: A narrative review. J. Cannabis Res. 2020, 2, 35. [Google Scholar] [CrossRef] [PubMed]






| Predicted | ||||
|---|---|---|---|---|
| Actual | Indica | Sativa | Sum | |
| Sativa | 238 | 1 | 239 | |
| Indica | 0 | 60 | 60 | |
| Sum | 238 | 61 | 299 | |
| Predicted | ||||||
|---|---|---|---|---|---|---|
| Actual | YGriega | OriginalAmnesia | AmnesiaHazeHypro | AmnesiaHaze | Sum | |
| YGriega | 54 | 0 | 0 | 0 | 54 | |
| OriginalAmnesia | 0 | 49 | 0 | 0 | 49 | |
| AmnesiaHazeHypro | 0 | 0 | 53 | 0 | 53 | |
| AmnesiaHaze | 1 | 0 | 0 | 58 | 59 | |
| Sum | 55 | 49 | 53 | 58 | 215 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos-Guerrero, L.; Montalvo, G.; Cosmi, M.; García-Ruiz, C.; Ortega-Ojeda, F.E. Classification of Various Marijuana Varieties by Raman Microscopy and Chemometrics. Toxics 2022, 10, 115. https://doi.org/10.3390/toxics10030115
Ramos-Guerrero L, Montalvo G, Cosmi M, García-Ruiz C, Ortega-Ojeda FE. Classification of Various Marijuana Varieties by Raman Microscopy and Chemometrics. Toxics. 2022; 10(3):115. https://doi.org/10.3390/toxics10030115
Chicago/Turabian StyleRamos-Guerrero, Luis, Gemma Montalvo, Marzia Cosmi, Carmen García-Ruiz, and Fernando E. Ortega-Ojeda. 2022. "Classification of Various Marijuana Varieties by Raman Microscopy and Chemometrics" Toxics 10, no. 3: 115. https://doi.org/10.3390/toxics10030115
APA StyleRamos-Guerrero, L., Montalvo, G., Cosmi, M., García-Ruiz, C., & Ortega-Ojeda, F. E. (2022). Classification of Various Marijuana Varieties by Raman Microscopy and Chemometrics. Toxics, 10(3), 115. https://doi.org/10.3390/toxics10030115