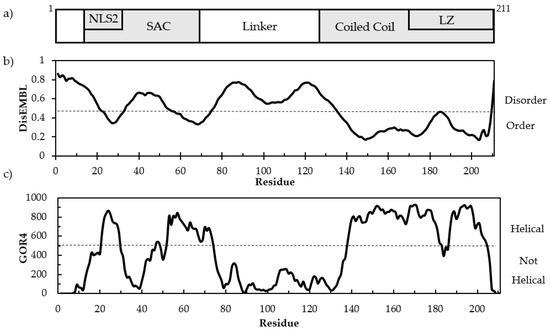
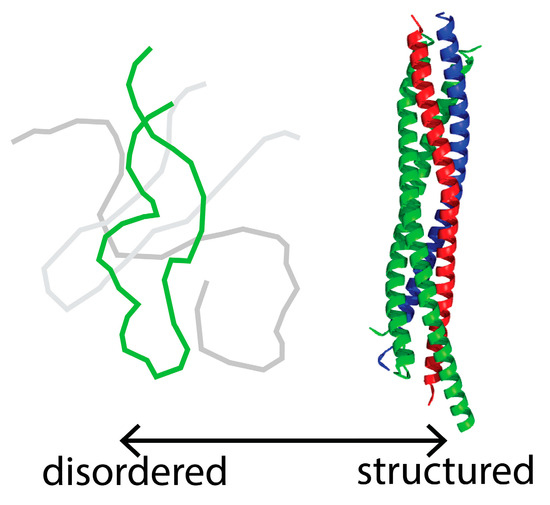
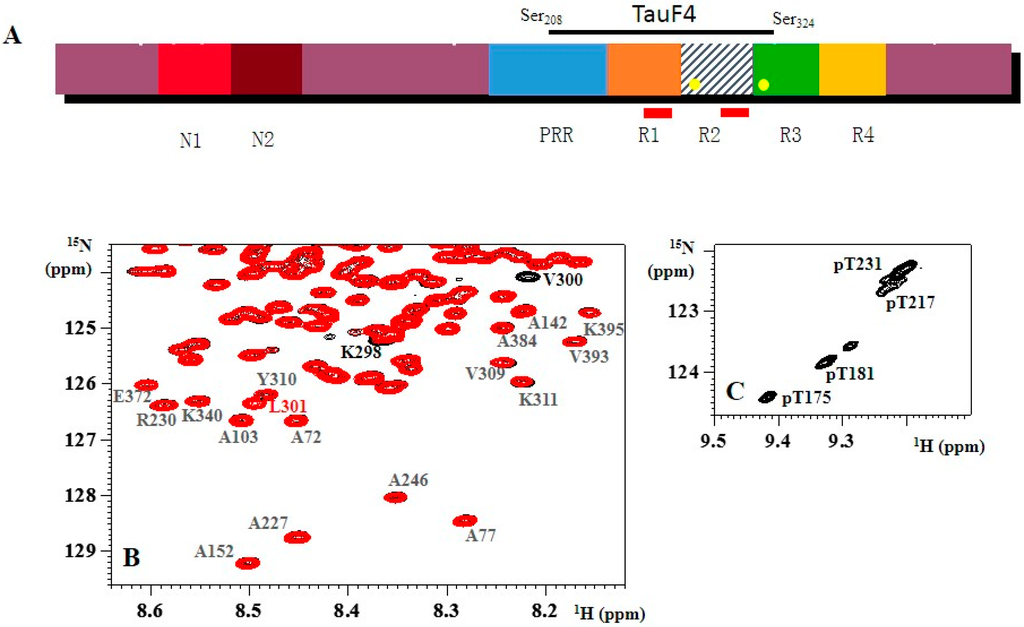
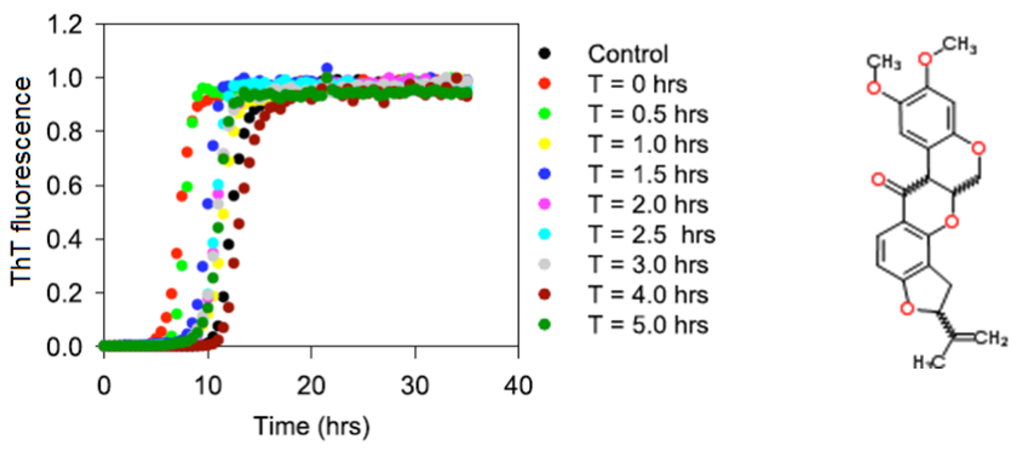
Intrinsically Disordered Proteins (Closed)
A topical collection in Biomolecules (ISSN 2218-273X). This collection belongs to the section "Cellular Biochemistry".
Viewed by 63610Editors
Interests: intrinsically disordered proteins; protein folding; protein misfolding; partially folded proteins; protein aggregation; protein structure; protein function; protein stability; protein biophysics; protein bioinformatics; conformational diseases; protein–ligand interactions; protein–protein interactions; liquid-liquid phase transitions
Special Issues, Collections and Topics in MDPI journals
Interests: tumor associated proteins; carbonic anhydrase; intrinsically disordered proteins; CAF-1 chemical biology; protein–protein interaction
Special Issues, Collections and Topics in MDPI journals
Interests: structural characterization of proteins; structural characterization of protein–ligand complexes; X ray crystallography; rational drug design; protein bioinformatics; carbonic anhydrase; cancer-related proteins
Special Issues, Collections and Topics in MDPI journals
Interests: computational chemistry; molecular dynamics simulations; modeling; protein-ligand docking; protein-protein docking; computer-aided drug design; binding free energy calculations; intrinsically disordered proteins; carbonic anhydrases; oligonucleotides; Alzheimer-related proteins; cancer-related proteins
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
As follows from the title, this Topical Collection “Intrinsically Disordered Proteins” aims to collect high-quality research articles, communications, and review articles in all the aspects related to the protein intrinsic disorder phenomenon. This Collection covers a wide range of topics, including structural manifestations of intrinsic disorder; functionality of intrinsically disordered proteins/regions; abundance of intrinsically disordered proteins; structure–function relationships for these proteins/regions; roles of intrinsic disorder in various biological processes; the intrinsic disorder-based mechanisms of regulation, recognition, and signaling; correlation between intrinsically disordered proteins and diseases; expression analysis of genes encoding intrinsically disordered proteins; description of experimental and computational techniques applicable for the analysis of structure and function of intrinsically disordered proteins.
The Topical Collection “Intrinsically Disordered Proteins” has several goals:
- To provide a platform devoted to protein intrinsic disorder that assembles top-quality papers on all aspects of this topic and promotes the field.
- To raise awareness about biological importance and abundance of intrinsically disordered proteins.
- To define and build a worldwide community of scientists interested in intrinsically disordered proteins, and to facilitate communication among them.
- To provide resources to enhance the effort of laboratories working on intrinsically disordered proteins.
I encourage you to share your data and thoughts in this broad field that clearly demonstrates the physiological and pathological importance of intrinsically disordered proteins.
Dr. Vladimir N. Uversky
Dr. Simona Maria Monti
Dr. Giuseppina De Simone
Dr. Emma Langella
Guest Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Biomolecules is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Planned Papers
The below list represents only planned manuscripts. Some of these manuscripts have not been received by the Editorial Office yet. Papers submitted to MDPI journals are subject to peer-review.
Title: To be determined
Authors: Michael A. Menze; et al.
Affiliation: Department of Biology, University of Louisville, Louisville, KY 40292, USA