Please note that, as of 18 July 2017, Microarrays has been renamed to High-Throughput and is now published here.
Journal Description
Microarrays
Microarrays
is an international peer-reviewed open access journal of microarray technology published quarterly online by MDPI. Note that from Volume 6, Issue 3, Microarrays has been renamed High-Throughput.
- Open Access - free for readers, with article processing charges (APC) paid by authors or their institutions.
- Rapid publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 28 days after submission; acceptance to publication is undertaken in 8 days (median values for papers published in this journal in 2016).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
- Microarrays has been recently renamed High-Throughput.
Latest Articles
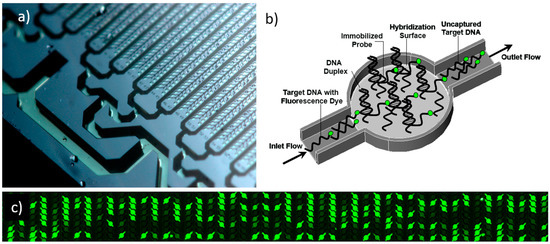
Modeling Hybridization Kinetics of Gene Probes in a DNA Biochip Using FEMLAB
Microarrays 2017, 6(2), 9; https://doi.org/10.3390/microarrays6020009 - 29 May 2017
Cited by 9
Abstract
Microfluidic DNA biochips capable of detecting specific DNA sequences are useful in medical diagnostics, drug discovery, food safety monitoring and agriculture. They are used as miniaturized platforms for analysis of nucleic acids-based biomarkers. Binding kinetics between immobilized single stranded DNA on the surface
[...] Read more.
Microfluidic DNA biochips capable of detecting specific DNA sequences are useful in medical diagnostics, drug discovery, food safety monitoring and agriculture. They are used as miniaturized platforms for analysis of nucleic acids-based biomarkers. Binding kinetics between immobilized single stranded DNA on the surface and its complementary strand present in the sample are of interest. To achieve optimal sensitivity with minimum sample size and rapid hybridization, ability to predict the kinetics of hybridization based on the thermodynamic characteristics of the probe is crucial. In this study, a computer aided numerical model for the design and optimization of a flow-through biochip was developed using a finite element technique packaged software tool (FEMLAB; package included in COMSOL Multiphysics) to simulate the transport of DNA through a microfluidic chamber to the reaction surface. The model accounts for fluid flow, convection and diffusion in the channel and on the reaction surface. Concentration, association rate constant, dissociation rate constant, recirculation flow rate, and temperature were key parameters affecting the rate of hybridization. The model predicted the kinetic profile and signal intensities of eighteen 20-mer probes targeting vancomycin resistance genes (VRGs). Predicted signal intensities and hybridization kinetics strongly correlated with experimental data in the biochip (R2 = 0.8131).
Full article
(This article belongs to the Special Issue Microfluidics Technology)
►
Show Figures
Open AccessArticle
Microarray Selection of Cooperative Peptides for Modulating Enzyme Activities
by
Jinglin Fu
Microarrays 2017, 6(2), 8; https://doi.org/10.3390/microarrays6020008 - 26 Apr 2017
Cited by 2
Abstract
Recently, peptide microarrays have been used to distinguish proteins, antibodies, viruses, and bacteria based on their binding to random sequence peptides. We reported on the use of peptide arrays to identify enzyme modulators that involve screening an array of 10,000 defined and addressable
[...] Read more.
Recently, peptide microarrays have been used to distinguish proteins, antibodies, viruses, and bacteria based on their binding to random sequence peptides. We reported on the use of peptide arrays to identify enzyme modulators that involve screening an array of 10,000 defined and addressable peptides on a microarray. Primary peptides were first selected to inhibit the enzyme at low μM concentrations. Then, new peptides were found to only bind strongly with the enzyme–inhibitor complex, but not the native enzyme. These new peptides served as secondary inhibitors that enhanced the inhibition of the enzyme together with the primary peptides. Without the primary peptides, the secondary effect peptides had little effect on the enzyme activity. Conversely, we also selected peptides that recovered the activities of inhibited enzyme–peptide complex. The selection of cooperative peptide pairs will provide a versatile toolkit for modulating enzyme functions, which may potentially be applied to drug discovery and biocatalysis.
Full article
(This article belongs to the Special Issue Microarrays for Drug Discovery and Development)
►▼
Show Figures
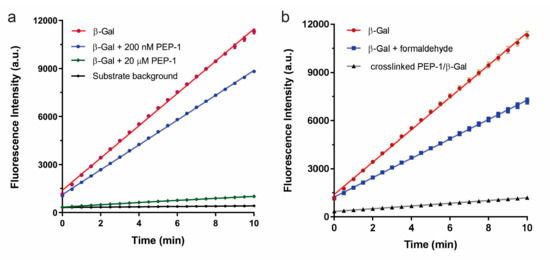
Figure 1
Open AccessReview
Avian and Mammalian Facilitative Glucose Transporters
by
Mary Shannon Byers, Christianna Howard and Xiaofei Wang
Microarrays 2017, 6(2), 7; https://doi.org/10.3390/microarrays6020007 - 5 Apr 2017
Cited by 23
Abstract
►▼
Show Figures
The GLUT members belong to a family of glucose transporter proteins that facilitate glucose transport across the cell membrane. The mammalian GLUT family consists of thirteen members (GLUTs 1–12 and H+-myo-inositol transporter (HMIT)). Humans have a recently duplicated GLUT member, GLUT14.
[...] Read more.
The GLUT members belong to a family of glucose transporter proteins that facilitate glucose transport across the cell membrane. The mammalian GLUT family consists of thirteen members (GLUTs 1–12 and H+-myo-inositol transporter (HMIT)). Humans have a recently duplicated GLUT member, GLUT14. Avians express the majority of GLUT members. The arrangement of multiple GLUTs across all somatic tissues signifies the important role of glucose across all organisms. Defects in glucose transport have been linked to metabolic disorders, insulin resistance and diabetes. Despite the essential importance of these transporters, our knowledge regarding GLUT members in avians is fragmented. It is clear that there are no chicken orthologs of mammalian GLUT4 and GLUT7. Our examination of GLUT members in the chicken revealed that some chicken GLUT members do not have corresponding orthologs in mammals. We review the information regarding GLUT orthologs and their function and expression in mammals and birds, with emphasis on chickens and humans.
Full article
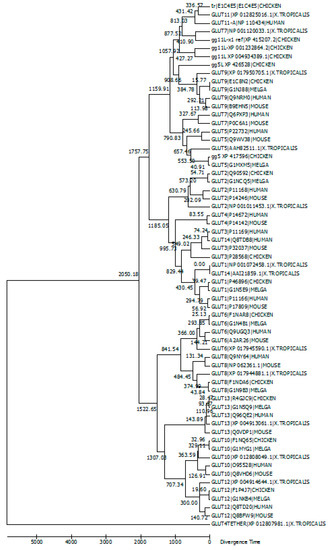
Figure 1
Open AccessReview
Use of a Pan–Genomic DNA Microarray in Determination of the Phylogenetic Relatedness among Cronobacter spp. and Its Use as a Data Mining Tool to Understand Cronobacter Biology
by
Ben D. Tall, Jayanthi Gangiredla, Christopher J. Grim, Isha R. Patel, Scott A. Jackson, Mark K. Mammel, Mahendra H. Kothary, Venugopal Sathyamoorthy, Laurenda Carter, Séamus Fanning, Carol Iversen, Franco Pagotto, Roger Stephan, Angelika Lehner, Jeffery Farber, Qiong Q. Yan and Gopal R. Gopinath
Microarrays 2017, 6(1), 6; https://doi.org/10.3390/microarrays6010006 - 4 Mar 2017
Cited by 5
Abstract
Cronobacter (previously known as Enterobacter sakazakii) is a genus of Gram-negative, facultatively anaerobic, oxidase-negative, catalase-positive, rod-shaped bacteria of the family Enterobacteriaceae. These organisms cause a variety of illnesses such as meningitis, necrotizing enterocolitis, and septicemia in neonates and infants, and urinary
[...] Read more.
Cronobacter (previously known as Enterobacter sakazakii) is a genus of Gram-negative, facultatively anaerobic, oxidase-negative, catalase-positive, rod-shaped bacteria of the family Enterobacteriaceae. These organisms cause a variety of illnesses such as meningitis, necrotizing enterocolitis, and septicemia in neonates and infants, and urinary tract, wound, abscesses or surgical site infections, septicemia, and pneumonia in adults. The total gene content of 379 strains of Cronobacter spp. and taxonomically-related isolates was determined using a recently reported DNA microarray. The Cronobacter microarray as a genotyping tool gives the global food safety community a rapid method to identify and capture the total genomic content of outbreak isolates for food safety, environmental, and clinical surveillance purposes. It was able to differentiate the seven Cronobacter species from one another and from non-Cronobacter species. The microarray was also able to cluster strains within each species into well-defined subgroups. These results also support previous studies on the phylogenic separation of species members of the genus and clearly highlight the evolutionary sequence divergence among each species of the genus compared to phylogenetically-related species. This review extends these studies and illustrates how the microarray can also be used as an investigational tool to mine genomic data sets from strains. Three case studies describing the use of the microarray are shown and include: (1) the determination of allelic differences among Cronobacter sakazakii strains possessing the virulence plasmid pESA3; (2) mining of malonate and myo-inositol alleles among subspecies of Cronobacter dublinensis strains to determine subspecies identity; and (3) lastly using the microarray to demonstrate sequence divergence and phylogenetic relatedness trends for 13 outer-membrane protein alleles among 240 Cronobacter and phylogenetically-related strains. The goal of this review is to describe microarrays as a robust tool for genomics research of this assorted and important genus, a criterion toward the development of future preventative measures to eliminate this foodborne pathogen from the global food supply.
Full article
(This article belongs to the Special Issue Microarrays for Food Safety and Quality)
►▼
Show Figures
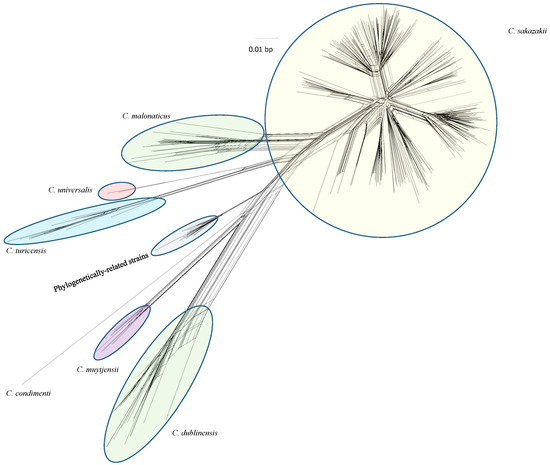
Figure 1
Open AccessArticle
A New Distribution Family for Microarray Data
by
Diana Mabel Kelmansky and Lila Ricci
Microarrays 2017, 6(1), 5; https://doi.org/10.3390/microarrays6010005 - 10 Feb 2017
Cited by 2
Abstract
The traditional approach with microarray data has been to apply transformations that approximately normalize them, with the drawback of losing the original scale. The alternative stand point taken here is to search for models that fit the data, characterized by the presence of
[...] Read more.
The traditional approach with microarray data has been to apply transformations that approximately normalize them, with the drawback of losing the original scale. The alternative stand point taken here is to search for models that fit the data, characterized by the presence of negative values, preserving their scale; one advantage of this strategy is that it facilitates a direct interpretation of the results. A new family of distributions named gpower-normal indexed by p∈R is introduced and it is proven that these variables become normal or truncated normal when a suitable gpower transformation is applied. Expressions are given for moments and quantiles, in terms of the truncated normal density. This new family can be used to model asymmetric data that include non-positive values, as required for microarray analysis. Moreover, it has been proven that the gpower-normal family is a special case of pseudo-dispersion models, inheriting all the good properties of these models, such as asymptotic normality for small variances. A combined maximum likelihood method is proposed to estimate the model parameters, and it is applied to microarray and contamination data. Rcodes are available from the authors upon request.
Full article
(This article belongs to the Special Issue Next Generation Microarray Bioinformatics)
►▼
Show Figures
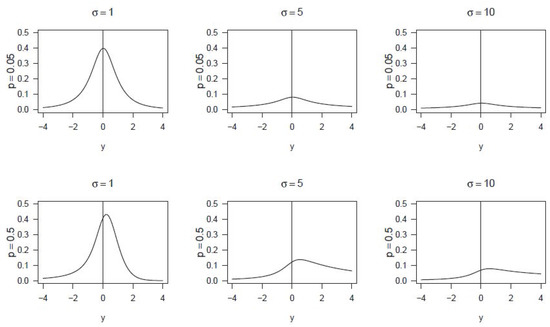
Figure 1
Open AccessReview
DNA Microarray‐Based Screening and Characterization of Traditional Chinese Medicine
by
Ryoiti Kiyama
Microarrays 2017, 6(1), 4; https://doi.org/10.3390/microarrays6010004 - 30 Jan 2017
Cited by 13
Abstract
The application of DNA microarray assay (DMA) has entered a new era owing to recent innovations in omics technologies. This review summarizes recent applications of DMA‐based gene expression profiling by focusing on the screening and characterizationof traditional Chinese medicine. First, herbs, mushrooms, and
[...] Read more.
The application of DNA microarray assay (DMA) has entered a new era owing to recent innovations in omics technologies. This review summarizes recent applications of DMA‐based gene expression profiling by focusing on the screening and characterizationof traditional Chinese medicine. First, herbs, mushrooms, and dietary plants analyzed by DMA along with their effective components and their biological/physiological effects are summarized and discussed by examining their comprehensive list and a list of representative effective chemicals. Second, the mechanisms of action of traditional Chinese medicine are summarized by examining the genes and pathways responsible for the action, the cell functions involved in the action, and the activities found by DMA (silent estrogens). Third, applications of DMA for traditional Chinese medicine are discussed by examining reported examples and new protocols for its use in quality control. Further innovations in the signaling pathway based evaluation of beneficial effects and the assessment of potential risks of traditional Chinese medicine are expected, just as are observed in other closely related fields, such as the therapeutic, environmental, nutritional, and pharmacological fields.
Full article
(This article belongs to the Special Issue Biochips)
►▼
Show Figures
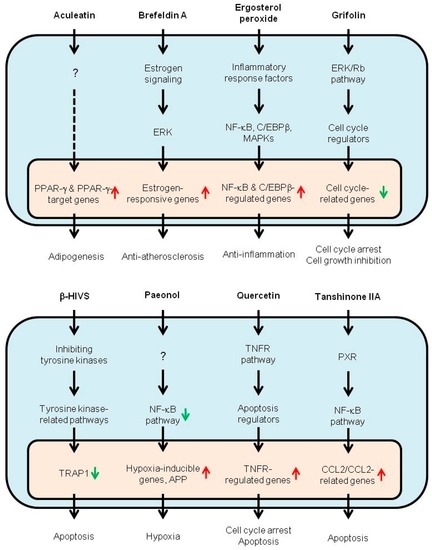
Figure 1
Open AccessReview
Microarray Technology Applied to Human Allergic Disease
by
Robert G. Hamilton
Microarrays 2017, 6(1), 3; https://doi.org/10.3390/microarrays6010003 - 28 Jan 2017
Cited by 13
Abstract
IgE antibodies serve as the gatekeeper for the release of mediators from sensitized (IgE positive) mast cells and basophils following a relevant allergen exposure which can lead to an immediate-type hypersensitivity (allergic) reaction. Purified recombinant and native allergens were combined in the 1990s
[...] Read more.
IgE antibodies serve as the gatekeeper for the release of mediators from sensitized (IgE positive) mast cells and basophils following a relevant allergen exposure which can lead to an immediate-type hypersensitivity (allergic) reaction. Purified recombinant and native allergens were combined in the 1990s with state of the art chip technology to establish the first microarray-based IgE antibody assay. Triplicate spots to over 100 allergenic molecules are immobilized on an amine-activated glass slide to form a single panel multi-allergosorbent assay. Human antibodies, typically of the IgE and IgG isotypes, specific for one or many allergens bind to their respective allergen(s) on the chip. Following removal of unbound serum proteins, bound IgE antibody is detected with a fluorophore-labeled anti-human IgE reagent. The fluorescent profile from the completed slide provides a sensitization profile of an allergic patient which can identify IgE antibodies that bind to structurally similar (cross-reactive) allergen families versus molecules that are unique to a single allergen specificity. Despite its ability to rapidly analyze many IgE antibody specificities in a single simple assay format, the chip-based microarray remains less analytically sensitive and quantitative than its singleplex assay counterpart (ImmunoCAP, Immulite). Microgram per mL quantities of allergen-specific IgG antibody can also complete with nanogram per mL quantities of specific IgE for limited allergen binding sites on the chip. Microarray assays, while not used in clinical immunology laboratories for routine patient IgE antibody testing, will remain an excellent research tool for defining sensitization profiles of populations in epidemiological studies.
Full article
(This article belongs to the Special Issue Microarrays in Immunology Research)
►▼
Show Figures
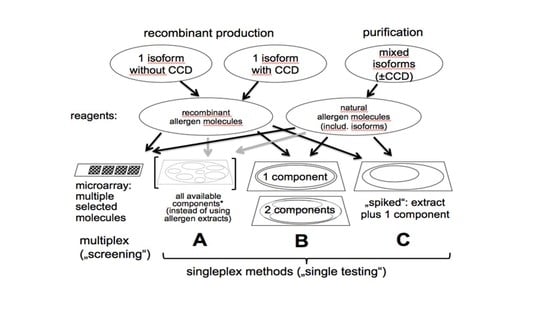
Graphical abstract
Open AccessEditorial
Acknowledgement to Reviewers of Microarrays in 2016
by
Microarrays Editorial Office
Microarrays 2017, 6(1), 2; https://doi.org/10.3390/microarrays6010002 - 11 Jan 2017
Abstract
The editors of Microarrays would like to express their sincere gratitude to the following reviewers for assessing manuscripts in 2016.[...]
Full article
Open AccessTechnical Note
Application of Lectin Array Technology for Biobetter Characterization: Its Correlation with FcγRIII Binding and ADCC
by
Markus Roucka, Klaus Zimmermann, Markus Fido and Andreas Nechansky
Microarrays 2017, 6(1), 1; https://doi.org/10.3390/microarrays6010001 - 24 Dec 2016
Cited by 9
Abstract
►▼
Show Figures
Lectin microarray technology was applied to compare the glycosylation pattern of the monoclonal antibody MB311 expressed in SP2.0 cells to an antibody-dependent cellular cytotoxic effector function (ADCC)-optimized variant (MB314). MB314 was generated by a plant expression system that uses genetically modified moss protoplasts
[...] Read more.
Lectin microarray technology was applied to compare the glycosylation pattern of the monoclonal antibody MB311 expressed in SP2.0 cells to an antibody-dependent cellular cytotoxic effector function (ADCC)-optimized variant (MB314). MB314 was generated by a plant expression system that uses genetically modified moss protoplasts (Physcomitrella patens) to generate a de-fucosylated version of MB311. In contrast to MB311, no or very low interactions of MB314 with lectins Aspergillus oryzae l-fucose (AOL), Pisum sativum agglutinin (PSA), Lens culinaris agglutinin (LCA), and Aleuria aurantia lectin (AAL) were observed. These lectins are specific for mono-/biantennary N-glycans containing a core fucose residue. Importantly, this fucose indicative lectin-binding pattern correlated with increased MB314 binding to CD16 (FcγRIII; receptor for the constant region of an antibody)—whose affinity is mediated through core fucosylation—and stronger ADCC. In summary, these results demonstrate that lectin microarrays are useful orthogonal methods during antibody development and for characterization.
Full article
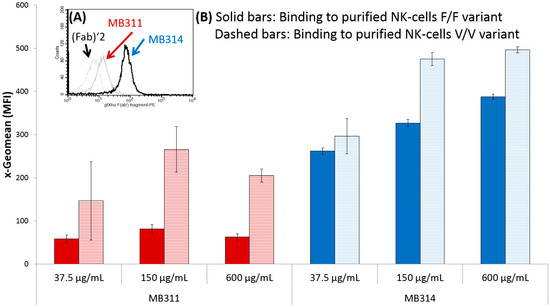
Figure 1
Open AccessArticle
Using miRNA-Analyzer for the Analysis of miRNA Data
by
Pietro Hiram Guzzi, Giuseppe Tradigo and Pierangelo Veltri
Microarrays 2016, 5(4), 29; https://doi.org/10.3390/microarrays5040029 - 15 Dec 2016
Abstract
MicroRNAs (miRNAs) are small biological molecules that play an important role during the mechanisms of protein formation. Recent findings have demonstrated that they act as both positive and negative regulators of protein formation. Thus, the investigation of miRNAs, i.e., the determination of their
[...] Read more.
MicroRNAs (miRNAs) are small biological molecules that play an important role during the mechanisms of protein formation. Recent findings have demonstrated that they act as both positive and negative regulators of protein formation. Thus, the investigation of miRNAs, i.e., the determination of their level of expression, has developed a huge interest in the scientific community. One of the leading technologies for extracting miRNA data from biological samples is the miRNA Affymetrix platform. It provides the quantification of the level of expression of the miRNA in a sample, thus enabling the accumulation of data and allowing the determination of relationships among miRNA, genes, and diseases. Unfortunately, there is a lack of a comprehensive platform able to provide all the functions needed for the extraction of information from miRNA data. We here present miRNA-Analyzer, a complete software tool providing primary functionalities for miRNA data analysis. The current version of miRNA-Analyzer wraps the Affymetrix QCTool for the preprocessing of binary data files, and then provides feature selection (the filtering by species and by the associated p-value of preprocessed files). Finally, preprocessed and filtered data are analyzed by the Multiple Experiment Viewer (T-MEV) and Short Time Series Expression Miner (STEM) tools, which are also wrapped into miRNA-Analyzer, thus providing a unique environment for miRNA data analysis. The tool offers a plug-in interface so it is easily extensible by adding other algorithms as plug-ins. Users may download the tool freely for academic use at https://sites.google.com/site/mirnaanalyserproject/d .
Full article
(This article belongs to the Special Issue Next Generation Microarray Bioinformatics)
►▼
Show Figures
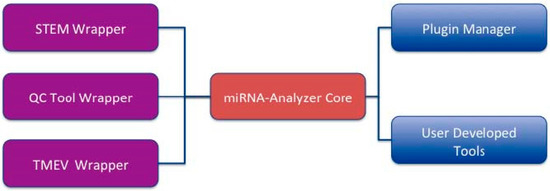
Figure 1
Open AccessArticle
Droplet Microarray Based on Superhydrophobic-Superhydrophilic Patterns for Single Cell Analysis
by
Gabriella E. Jogia, Tina Tronser, Anna A. Popova and Pavel A. Levkin
Microarrays 2016, 5(4), 28; https://doi.org/10.3390/microarrays5040028 - 9 Dec 2016
Cited by 28
Abstract
Single-cell analysis provides fundamental information on individual cell response to different environmental cues and is a growing interest in cancer and stem cell research. However, current existing methods are still facing challenges in performing such analysis in a high-throughput manner whilst being cost-effective.
[...] Read more.
Single-cell analysis provides fundamental information on individual cell response to different environmental cues and is a growing interest in cancer and stem cell research. However, current existing methods are still facing challenges in performing such analysis in a high-throughput manner whilst being cost-effective. Here we established the Droplet Microarray (DMA) as a miniaturized screening platform for high-throughput single-cell analysis. Using the method of limited dilution and varying cell density and seeding time, we optimized the distribution of single cells on the DMA. We established culturing conditions for single cells in individual droplets on DMA obtaining the survival of nearly 100% of single cells and doubling time of single cells comparable with that of cells cultured in bulk cell population using conventional methods. Our results demonstrate that the DMA is a suitable platform for single-cell analysis, which carries a number of advantages compared with existing technologies allowing for treatment, staining and spot-to-spot analysis of single cells over time using conventional analysis methods such as microscopy.
Full article
(This article belongs to the Special Issue Cell-Based Microarrays)
►▼
Show Figures
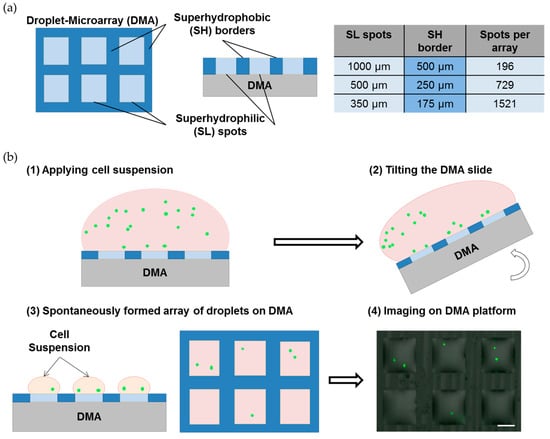
Figure 1
Open AccessEditorial
SNP Arrays
by
Jari Louhelainen
Microarrays 2016, 5(4), 27; https://doi.org/10.3390/microarrays5040027 - 25 Oct 2016
Cited by 6
Abstract
The papers published in this Special Issue “SNP arrays” (Single Nucleotide Polymorphism Arrays) focus on several perspectives associated with arrays of this type. The range of papers vary from a case report to reviews, thereby targeting wider audiences working in this field. The
[...] Read more.
The papers published in this Special Issue “SNP arrays” (Single Nucleotide Polymorphism Arrays) focus on several perspectives associated with arrays of this type. The range of papers vary from a case report to reviews, thereby targeting wider audiences working in this field. The research focus of SNP arrays is often human cancers but this Issue expands that focus to include areas such as rare conditions, animal breeding and bioinformatics tools. Given the limited scope, the spectrum of papers is nothing short of remarkable and even from a technical point of view these papers will contribute to the field at a general level. Three of the papers published in this Special Issue focus on the use of various SNP array approaches in the analysis of three different cancer types. Two of the papers concentrate on two very different rare conditions, applying the SNP arrays slightly differently. Finally, two other papers evaluate the use of the SNP arrays in the context of genetic analysis of livestock. The findings reported in these papers help to close gaps in the current literature and also to give guidelines for future applications of SNP arrays.
Full article
(This article belongs to the Special Issue SNP Array)
Open AccessEditorial
Computational Modeling and Analysis of Microarray Data: New Horizons
by
Heather J. Ruskin
Microarrays 2016, 5(4), 26; https://doi.org/10.3390/microarrays5040026 - 21 Oct 2016
Cited by 5
Abstract
High-throughput microarray technologies have long been a source of data for a wide range of biomedical investigations. Over the decades, variants have been developed and sophistication of measurements has improved, with generated data providing both valuable insight and considerable analytical challenge. The cost-effectiveness
[...] Read more.
High-throughput microarray technologies have long been a source of data for a wide range of biomedical investigations. Over the decades, variants have been developed and sophistication of measurements has improved, with generated data providing both valuable insight and considerable analytical challenge. The cost-effectiveness of microarrays, as well as their fundamental applicability, made them a first choice for much early genomic research and efforts to improve accessibility, quality and interpretation have continued unabated. In recent years, however, the emergence of new generations of sequencing methods and, importantly, reduction of costs, has seen a preferred shift in much genomic research to the use of sequence data, both less ‘noisy’ and, arguably, with species information more directly targeted and easily interpreted. Nevertheless, new microarray data are still being generated and, together with their considerable legacy, can offer a complementary perspective on biological systems and disease pathogenesis. The challenge now is to exploit novel methods for enhancing and combining these data with those generated by alternative high-throughput techniques, such as sequencing, to provide added value. Augmentation and integration of microarray data and the new horizons this opens up, provide the theme for the papers in this Special Issue.
Full article
(This article belongs to the Special Issue Computational Modeling and Analysis of Microarray Data: New Horizons)
Open AccessArticle
Automated and Multiplexed Soft Lithography for the Production of Low-Density DNA Microarrays
by
Julie Fredonnet, Julie Foncy, Jean-Christophe Cau, Childérick Séverac, Jean Marie François and Emmanuelle Trévisiol
Microarrays 2016, 5(4), 25; https://doi.org/10.3390/microarrays5040025 - 26 Sep 2016
Cited by 4
Abstract
►▼
Show Figures
Microarrays are established research tools for genotyping, expression profiling, or molecular diagnostics in which DNA molecules are precisely addressed to the surface of a solid support. This study assesses the fabrication of low-density oligonucleotide arrays using an automated microcontact printing device, the InnoStamp
[...] Read more.
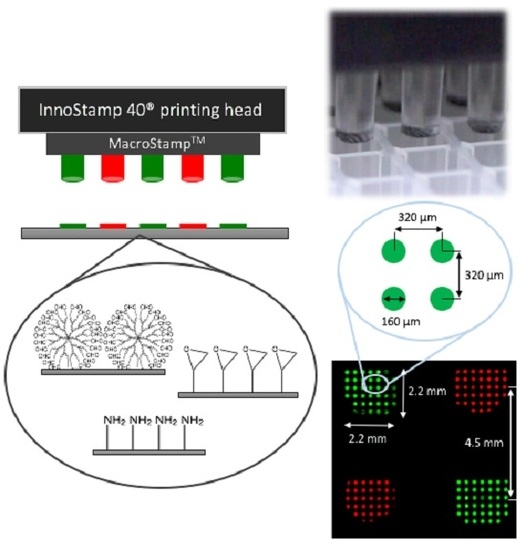
Microarrays are established research tools for genotyping, expression profiling, or molecular diagnostics in which DNA molecules are precisely addressed to the surface of a solid support. This study assesses the fabrication of low-density oligonucleotide arrays using an automated microcontact printing device, the InnoStamp 40®. This automate allows a multiplexed deposition of oligoprobes on a functionalized surface by the use of a MacroStampTM bearing 64 individual pillars each mounted with 50 circular micropatterns (spots) of 160 µm diameter at 320 µm pitch. Reliability and reuse of the MacroStampTM were shown to be fast and robust by a simple washing step in 96% ethanol. The low-density microarrays printed on either epoxysilane or dendrimer-functionalized slides (DendriSlides) showed excellent hybridization response with complementary sequences at unusual low probe and target concentrations, since the actual probe density immobilized by this technology was at least 10-fold lower than with the conventional mechanical spotting. In addition, we found a comparable hybridization response in terms of fluorescence intensity between spotted and printed oligoarrays with a 1 nM complementary target by using a 50-fold lower probe concentration to produce the oligoarrays by the microcontact printing method. Taken together, our results lend support to the potential development of this multiplexed microcontact printing technology employing soft lithography as an alternative, cost-competitive tool for fabrication of low-density DNA microarrays.
Full article
Graphical abstract
Open AccessArticle
OSAnalyzer: A Bioinformatics Tool for the Analysis of Gene Polymorphisms Enriched with Clinical Outcomes
by
Giuseppe Agapito, Cirino Botta, Pietro Hiram Guzzi, Mariamena Arbitrio, Maria Teresa Di Martino, Pierfrancesco Tassone, Pierosandro Tagliaferri and Mario Cannataro
Microarrays 2016, 5(4), 24; https://doi.org/10.3390/microarrays5040024 - 23 Sep 2016
Cited by 7
Abstract
Background: The identification of biomarkers for the estimation of cancer patients’ survival is a crucial problem in modern oncology. Recently, the Affymetrix DMET (Drug Metabolizing Enzymes and Transporters) microarray platform has offered the possibility to determine the ADME (absorption, distribution, metabolism, and excretion)
[...] Read more.
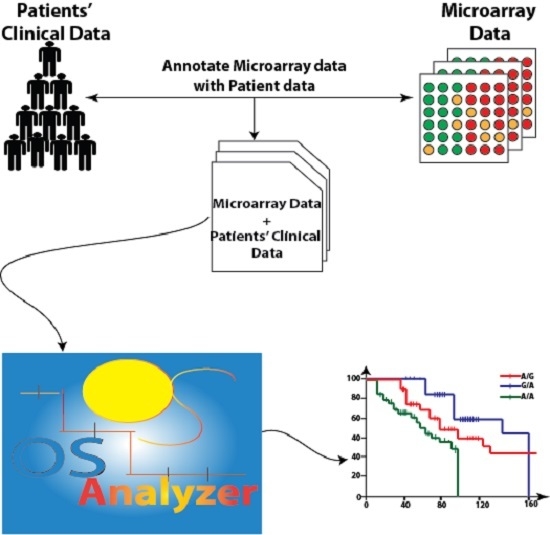
Background: The identification of biomarkers for the estimation of cancer patients’ survival is a crucial problem in modern oncology. Recently, the Affymetrix DMET (Drug Metabolizing Enzymes and Transporters) microarray platform has offered the possibility to determine the ADME (absorption, distribution, metabolism, and excretion) gene variants of a patient and to correlate them with drug-dependent adverse events. Therefore, the analysis of survival distribution of patients starting from their profile obtained using DMET data may reveal important information to clinicians about possible correlations among drug response, survival rate, and gene variants. Methods: In order to provide support to this analysis we developed OSAnalyzer, a software tool able to compute the overall survival (OS) and progression-free survival (PFS) of cancer patients and evaluate their association with ADME gene variants. Results: The tool is able to perform an automatic analysis of DMET data enriched with survival events. Moreover, results are ranked according to statistical significance obtained by comparing the area under the curves that is computed by using the log-rank test, allowing a quick and easy analysis and visualization of high-throughput data. Conclusions: Finally, we present a case study to highlight the usefulness of OSAnalyzer when analyzing a large cohort of patients.
Full article
(This article belongs to the Special Issue Next Generation Microarray Bioinformatics)
►▼
Show Figures
Graphical abstract
Open AccessFeature PaperArticle
Microarray Data Processing Techniques for Genome-Scale Network Inference from Large Public Repositories
by
Sriram Chockalingam, Maneesha Aluru and Srinivas Aluru
Microarrays 2016, 5(3), 23; https://doi.org/10.3390/microarrays5030023 - 19 Sep 2016
Cited by 11
Abstract
Pre-processing of microarray data is a well-studied problem. Furthermore, all popular platforms come with their own recommended best practices for differential analysis of genes. However, for genome-scale network inference using microarray data collected from large public repositories, these methods filter out a considerable
[...] Read more.
Pre-processing of microarray data is a well-studied problem. Furthermore, all popular platforms come with their own recommended best practices for differential analysis of genes. However, for genome-scale network inference using microarray data collected from large public repositories, these methods filter out a considerable number of genes. This is primarily due to the effects of aggregating a diverse array of experiments with different technical and biological scenarios. Here we introduce a pre-processing pipeline suitable for inferring genome-scale gene networks from large microarray datasets. We show that partitioning of the available microarray datasets according to biological relevance into tissue- and process-specific categories significantly extends the limits of downstream network construction. We demonstrate the effectiveness of our pre-processing pipeline by inferring genome-scale networks for the model plant Arabidopsis thaliana using two different construction methods and a collection of 11,760 Affymetrix ATH1 microarray chips. Our pre-processing pipeline and the datasets used in this paper are made available at http://alurulab.cc.gatech.edu/microarray-pp .
Full article
(This article belongs to the Special Issue Next Generation Microarray Bioinformatics)
►▼
Show Figures

Graphical abstract
Open AccessArticle
Serology in the Digital Age: Using Long Synthetic Peptides Created from Nucleic Acid Sequences as Antigens in Microarrays
by
Muhammad Rizwan, Bengt Rönnberg, Maksims Cistjakovs, Åke Lundkvist, Rudiger Pipkorn and Jonas Blomberg
Microarrays 2016, 5(3), 22; https://doi.org/10.3390/microarrays5030022 - 10 Aug 2016
Cited by 6
Abstract
Background: Antibodies to microbes, or to autoantigens, are important markers of disease. Antibody detection (serology) can reveal both past and recent infections. There is a great need for development of rational ways of detecting and quantifying antibodies, both for humans and animals. Traditionally,
[...] Read more.
Background: Antibodies to microbes, or to autoantigens, are important markers of disease. Antibody detection (serology) can reveal both past and recent infections. There is a great need for development of rational ways of detecting and quantifying antibodies, both for humans and animals. Traditionally, serology using synthetic antigens covers linear epitopes using up to 30 amino acid peptides. Methods: We here report that peptides of 100 amino acids or longer (“megapeptides”), designed and synthesized for optimal serological performance, can successfully be used as detection antigens in a suspension multiplex immunoassay (SMIA). Megapeptides can quickly be created just from pathogen sequences. A combination of rational sequencing and bioinformatic routines for definition of diagnostically-relevant antigens can, thus, rapidly yield efficient serological diagnostic tools for an emerging infectious pathogen. Results: We designed megapeptides using bioinformatics and viral genome sequences. These long peptides were tested as antigens for the presence of antibodies in human serum to the filo-, herpes-, and polyoma virus families in a multiplex microarray system. All of these virus families contain recently discovered or emerging infectious viruses. Conclusion: Long synthetic peptides can be useful as serological diagnostic antigens, serving as biomarkers, in suspension microarrays.
Full article
(This article belongs to the Special Issue High-Throughput Microarray for Protein Biomarker Discovery)
►▼
Show Figures
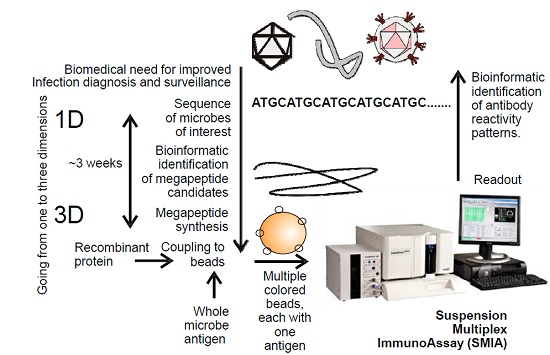
Graphical abstract
Open AccessArticle
A Combinatorial Protein Microarray for Probing Materials Interaction with Pancreatic Islet Cell Populations
by
Bahman Delalat, Darling M. Rojas-Canales, Soraya Rasi Ghaemi, Michaela Waibel, Frances J. Harding, Daniella Penko, Christopher J. Drogemuller, Thomas Loudovaris, Patrick T. H. Coates and Nicolas H. Voelcker
Microarrays 2016, 5(3), 21; https://doi.org/10.3390/microarrays5030021 - 10 Aug 2016
Cited by 5
Abstract
Pancreatic islet transplantation has become a recognized therapy for insulin-dependent diabetes mellitus. During isolation from pancreatic tissue, the islet microenvironment is disrupted. The extracellular matrix (ECM) within this space not only provides structural support, but also actively signals to regulate islet survival and
[...] Read more.
Pancreatic islet transplantation has become a recognized therapy for insulin-dependent diabetes mellitus. During isolation from pancreatic tissue, the islet microenvironment is disrupted. The extracellular matrix (ECM) within this space not only provides structural support, but also actively signals to regulate islet survival and function. In addition, the ECM is responsible for growth factor presentation and sequestration. By designing biomaterials that recapture elements of the native islet environment, losses in islet function and number can potentially be reduced. Cell microarrays are a high throughput screening tool able to recreate a multitude of cellular niches on a single chip. Here, we present a screening methodology for identifying components that might promote islet survival. Automated fluorescence microscopy is used to rapidly identify islet derived cell interaction with ECM proteins and immobilized growth factors printed on arrays. MIN6 mouse insulinoma cells, mouse islets and, finally, human islets are progressively screened. We demonstrate the capability of the platform to identify ECM and growth factor protein candidates that support islet viability and function and reveal synergies in cell response.
Full article
(This article belongs to the Special Issue Cell-Based Microarrays)
►▼
Show Figures
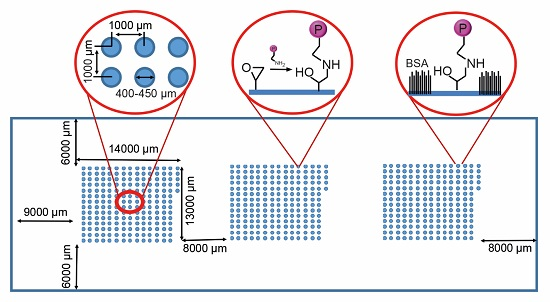
Graphical abstract
Open AccessReview
The Potentials and Pitfalls of Microarrays in Neglected Tropical Diseases: A Focus on Human Filarial Infections
by
Alexander Kwarteng and Samuel Terkper Ahuno
Microarrays 2016, 5(3), 20; https://doi.org/10.3390/microarrays5030020 - 2 Aug 2016
Cited by 1
Abstract
Data obtained from expression microarrays enables deeper understanding of the molecular signatures of infectious diseases. It provides rapid and accurate information on how infections affect the clustering of gene expression profiles, pathways and networks that are transcriptionally active during various infection states compared
[...] Read more.
Data obtained from expression microarrays enables deeper understanding of the molecular signatures of infectious diseases. It provides rapid and accurate information on how infections affect the clustering of gene expression profiles, pathways and networks that are transcriptionally active during various infection states compared to conventional diagnostic methods, which primarily focus on single genes or proteins. Thus, microarray technologies offer advantages in understanding host-parasite interactions associated with filarial infections. More importantly, the use of these technologies can aid diagnostics and helps translate current genomic research into effective treatment and interventions for filarial infections. Studying immune responses via microarray following infection can yield insight into genetic pathways and networks that can have a profound influence on the development of anti-parasitic vaccines.
Full article
(This article belongs to the Special Issue Microarrays in Immunology Research)
►▼
Show Figures
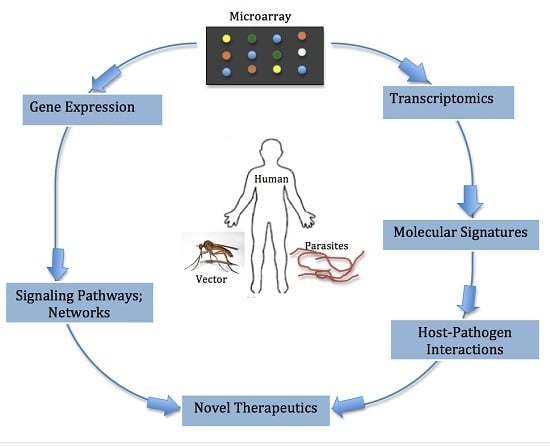
Graphical abstract
Open AccessArticle
Protein Profiling Gastric Cancer and Neighboring Control Tissues Using High-Content Antibody Microarrays
by
Martin Sill, Christoph Schröder, Ying Shen, Aseel Marzoq, Radovan Komel, Jörg D. Hoheisel, Henrik Nienhüser, Thomas Schmidt and Damjana Kastelic
Microarrays 2016, 5(3), 19; https://doi.org/10.3390/microarrays5030019 - 8 Jul 2016
Cited by 5
Abstract
In this study, protein profiling was performed on gastric cancer tissue samples in order to identify proteins that could be utilized for an effective diagnosis of this highly heterogeneous disease and as targets for therapeutic approaches. To this end, 16 pairs of postoperative
[...] Read more.
In this study, protein profiling was performed on gastric cancer tissue samples in order to identify proteins that could be utilized for an effective diagnosis of this highly heterogeneous disease and as targets for therapeutic approaches. To this end, 16 pairs of postoperative gastric adenocarcinomas and adjacent non-cancerous control tissues were analyzed on microarrays that contain 813 antibodies targeting 724 proteins. Only 17 proteins were found to be differentially regulated, with much fewer molecules than the numbers usually identified in studies comparing tumor to healthy control tissues. Insulin-like growth factor-binding protein 7 (IGFBP7), S100 calcium binding protein A9 (S100A9), interleukin-10 (IL‐10) and mucin 6 (MUC6) exhibited the most profound variations. For an evaluation of the proteins’ capacity for discriminating gastric cancer, a Receiver Operating Characteristic curve analysis was performed, yielding an accuracy (area under the curve) value of 89.2% for distinguishing tumor from non-tumorous tissue. For confirmation, immunohistological analyses were done on tissue slices prepared from another cohort of patients with gastric cancer. The utility of the 17 marker proteins, and particularly the four molecules with the highest specificity for gastric adenocarcinoma, is discussed for them to act as candidates for diagnosis, even in serum, and targets for therapeutic approaches.
Full article
(This article belongs to the Special Issue Antibody Microarrays in Clinical Proteomics)
►▼
Show Figures
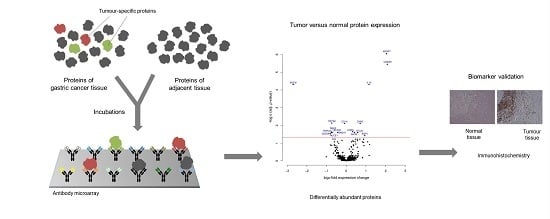
Graphical abstract
Highly Accessed Articles
Latest Books
E-Mail Alert
News
Topics