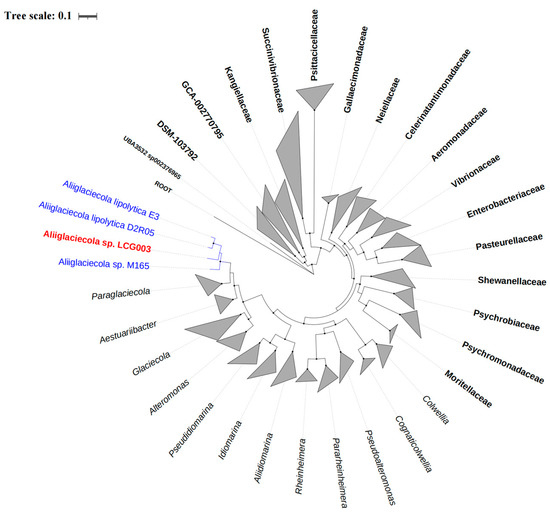
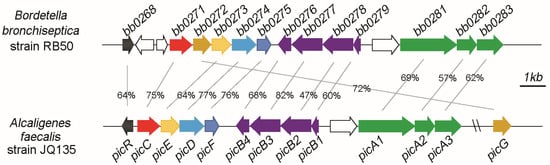
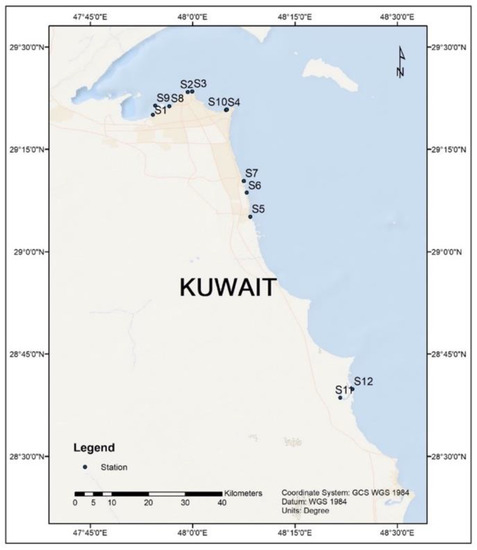
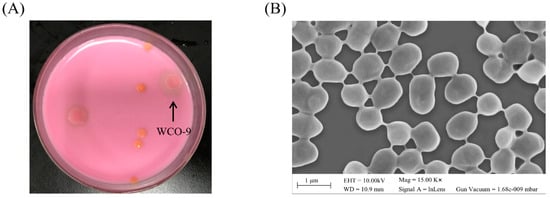
Biodegradation and Environmental Microbiomes
A topical collection in Microorganisms (ISSN 2076-2607). This collection belongs to the section "Environmental Microbiology".
Viewed by 24030Editors
Interests: environmental microbiology; bioremediation; synthetic biology; biodegradation; biotransformation
Special Issues, Collections and Topics in MDPI journals
Interests: biodegradation; bioremediation; catabolism; dehalogenation; biotransformation
Special Issues, Collections and Topics in MDPI journals
Interests: petroleum degradation; microbial enhanced oil recovery; xenobiotic compound degradation; microbial culture; wastewater treatment
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
This Topic Collection is the continuation of our previous Special Issue "Biodegradation and Environmental Microbiomes".
The Earth is unique, and we human beings rely on its air, water, and land. Industrialization and human activities have improved our daily life at the cost of nature resources and environmental quality. Air pollution, water eutrophication, and land deterioration challenge our sustainable development, and new technologies are needed to address these challenges. Biodegradation and bioremediation are promising technologies that can return humanity to a sustainable development. Microbe, or microbiome (the sum of all microbes in a defined environment) is the main driving force for biodegradation and bioremediation. This Topic Collection will cover new understandings of 1) what the nature and degree of air, water, and land pollution are, 2) how pollutants are degraded by natural or engineered microbes/microbiomes, and 3) successful large-scale implementation of biotechnologies for an improved environment. Both research articles and reviews are welcome.
Prof. Dr. Hongzhi Tang
Prof. Dr. Jiandong Jiang
Prof. Dr. Xiaolei Wu
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Microorganisms is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Related Special Issue
- Biodegradation and Environmental Microbiomes in Microorganisms (18 articles)