Exploration and Application of Useful Agricultural Genes
A topical collection in Plants (ISSN 2223-7747). This collection belongs to the section "Plant Genetics, Genomics and Biotechnology".
Viewed by 33689Editor
Interests: development of molecular markers related to pest resistance and search for candidate genes; host resistance (Host-plant resistance) mechanism of resistance-related research and object selection; pest-specific molecular markers development; population genetic analysis; search and use of agricultural useful genes using high-density genetic maps based on NGS technology
Topical Collection Information
Dear Colleagues,
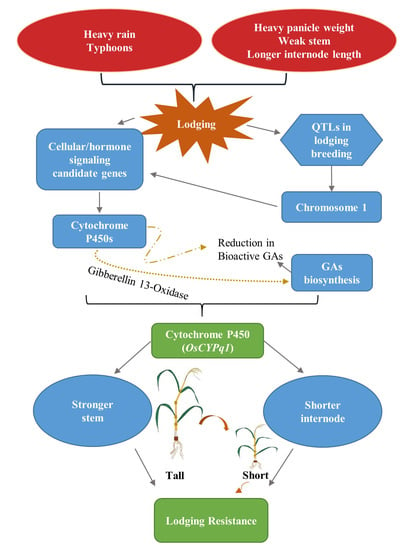
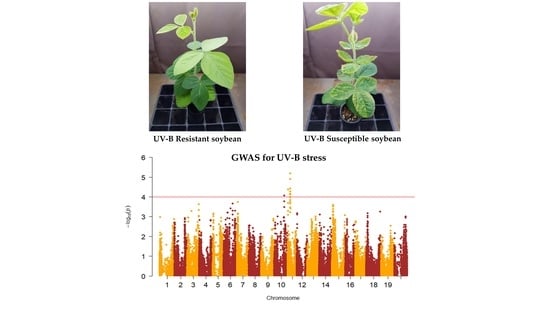
In the past decades, many studies on the search and use of agricultural useful genes have been actively conducted in various crops. As a result, researchers have found many useful genes involved in the desired agronomic traits through methods such as QTL mapping, genome-wide association study (GWAS), and transcriptome analysis. In addition, various types of molecular markers have been developed for practical use of the identified genes.
This Topical Collection welcomes original research papers as well as review articles related to recent molecular breeding studies involving identification of agronomically useful genes or QTL, development of molecular markers tightly linked to agronomic traits, and application of molecular markers in crop improvement.
Prof. Dr. Tae-Hwan Jun
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Plants is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2700 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- Agronomic trait
- QTL mapping
- Genome-wide association studies (GWAS)
- Transcriptome analysis
- Molecular markers
- Molecular plant breeding