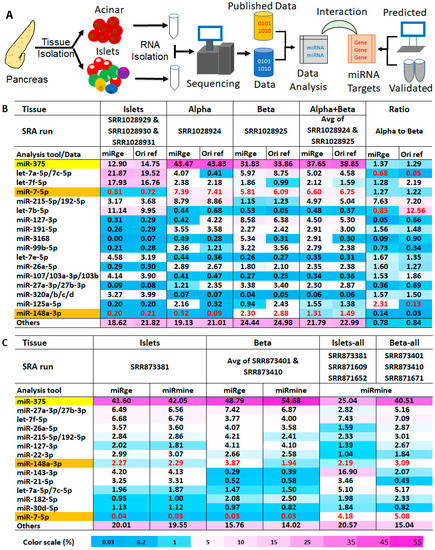
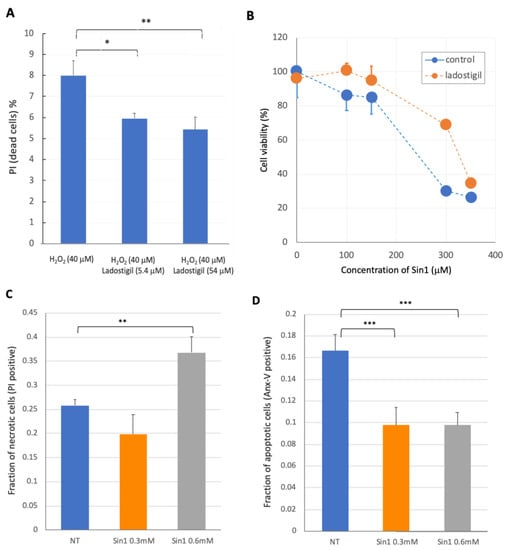
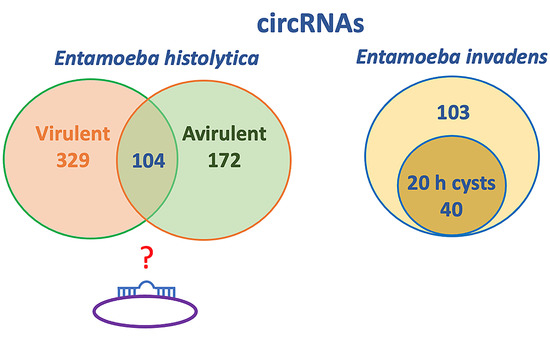
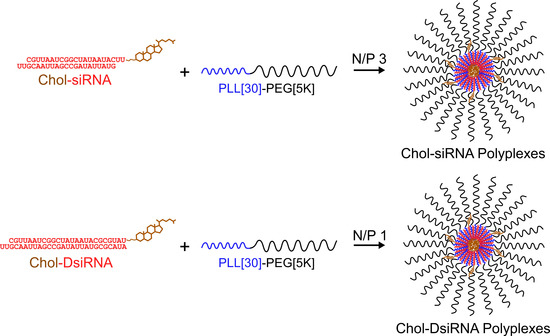
Feature Papers in Non-Coding RNA
A topical collection in Non-Coding RNA (ISSN 2311-553X). This collection belongs to the section "Computational Biology".
Viewed by 162016Editor
Interests: non-coding RNA; microRNA; cancer; metastasis
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
This Topical Collection “Feature Papers in Non-coding RNA” aims to collect high-quality research articles, review articles, and communications in all the fields of non-coding RNA research and their regulatory roles. Since the aim of this Topical Collection is to illustrate, through selected works, frontier research in noncoding RNA, we encourage Editorial Board Members of Non-coding RNA to contribute papers reflecting the latest progress in their research field, or relevant experts and colleagues to do so. Please kindly note that only invited papers can be published online once accepted in this collection.
Topics include, without being limited to, the following:
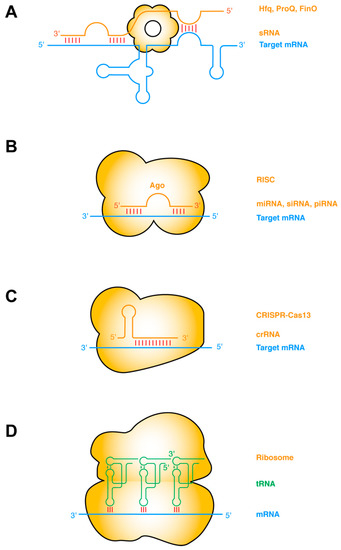
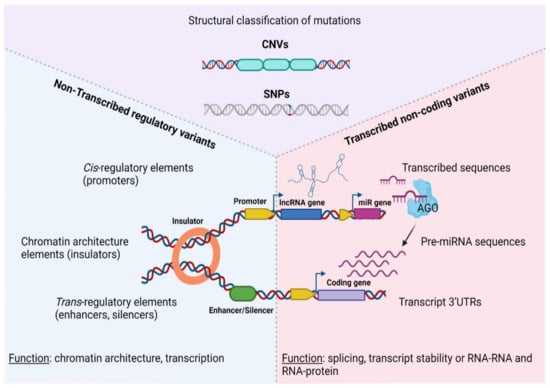
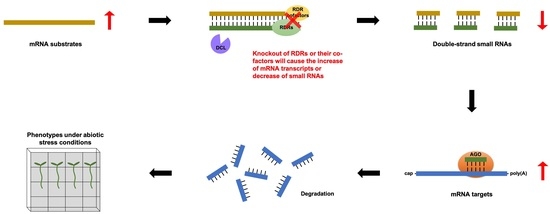
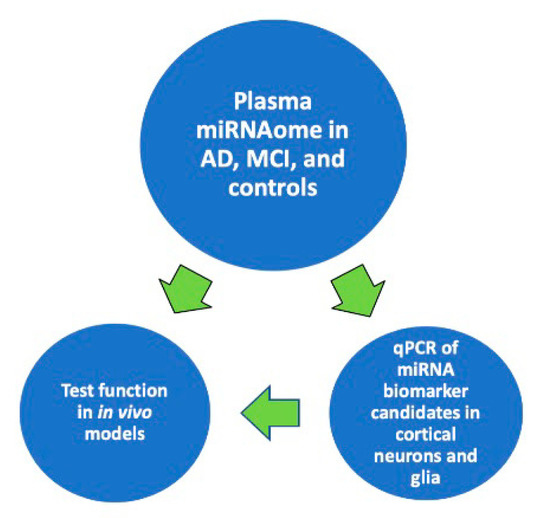
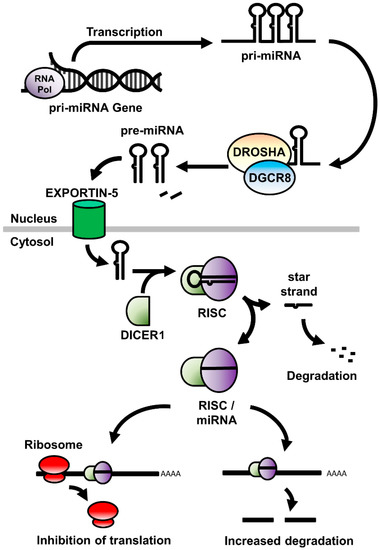
- Functional studies dealing with the identification, structure–function relationships, or biological activity of miRNAs, siRNAs, piRNAs, tRNAs, long noncoding RNAs, and other classes of RNAs;
- Analysis of RNA processing, RNA binding proteins, RNA signaling, and RNA interaction pathways;
- RNA analyses, informatics, tools, and technologies;
- Translational studies involving long and short noncoding RNAs.
Prof. Dr. Georges A. Calin
Guest Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Non-Coding RNA is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 1800 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Planned Papers
The below list represents only planned manuscripts. Some of these manuscripts have not been received by the Editorial Office yet. Papers submitted to MDPI journals are subject to peer-review.
Title: Circulating RNAs associated with the metabolic syndrome
Authors: Louise Torp Dalgaard
Affiliation: Eukaryot Cellebiologi, The Department of Science, Systems and Models, Roskilde University, Universitetsvej 1, 18.2, DK-4000, Roskilde, Denmark
Title: Bioinformatic analyses of conserved microRNAs-target gene networks between the human left ventricle and peripheral plasma could reveal new functional biomarkers of ischaemic heart disease in diabetic patients
Authors: Maryam Anwar1, Junyi Liu1, Ibrahim Nasser1, Gianni Angelini1,2, Prakash Punjabi1, Kerrie Ford, Aranzazu Chamorro-Jorganes1, Costanza Emanueli1, 2
Affiliation: 1National Heart and Lung Institute, Imperial College London, London, UK 2Bristol Heart Institute, University of Bristol, Bristol, UK
Title: ncRNAs and prostate cancer from a diagnostic point of view
Authors: Barbara Pardini
Affiliation: Italian Institute for Genomic Medicine - IIGM, c/o FPO IRCCS, 10060 Candiolo (To), Italy
Title: MiR-294 and miR-410 negatively regulates Tnfa, arginine transporters Cat1/2, and Nos2 mRNAs in macrophages infected with Leishmania amazonensis
Authors: Stephanie Maia Acuña, Jonathan Miguel Zanatta, Camilla de Almeida Bento, Lucile Maria Floeter-Winter, Sandra Marcia Muxel.
Affiliation: Departamento de Fisiologia, Instituto de Biociências, Universidade de São Paulo, São Paulo, SP, Brazil.
Abstract: MicroRNAs are small non-coding RNA that regulate post-transcriptionally several physiological processes, including immune responses. Murine macrophages infected with Leishmania amazonensis present crucial inflammatory response changes and metabolism accompanied by miRNA profile shifting. In this context, L-arginine is one of the mechanisms that modulate the inflammatory response through Nitric Oxide Synthase 2 (Nos2), producing NO, or arginase 1 (Arg1), providing fueling polyamine formation. This work aims to evaluate the expression and function of miR-294 and miR-410 during C57BL/6 bone marrow-derived macrophages early infection with L. amazonensis. We observed that after 4 and 24h of infection, miR-294 and miR-410 levels were upregulated, correlating with the decreased levels of mRNAs coding for tumor necrosis factor-alpha (Tnfa), Nos2, and cationic amino acid transporters 1 and 2 (Cat1 and Cat2). The functional inhibition of miR-294 led to increased levels of Tnfa, Nos2, and Cat2. Likewise, miR-410 inhibition increased Cat1 levels. Moreover, Nos2-/- infected macrophages showed decreased levels of Tnfa, but the miR-294 level remained the same. Also, miRNA inhibition reduced the parasite’s infectivity. Altogether, these data suggest that the microRNAs miR-294 and miR-410 play a role in the macrophage inflammatory response against L. amazonensis, contributing to the infection outcome definition.
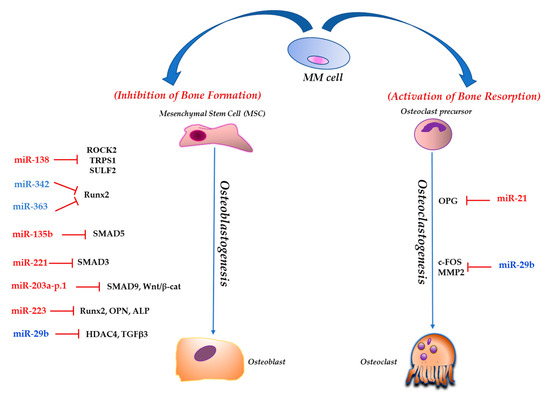
Title: non coding RNA impacting on mitochondrial functions
Authors: Nicola Amodio
Affiliation: Department of Experimental and Clinical Medicine, Magna Graecia University of Catanzaro, 88100 Catanzaro, Italy
Title: Evolution of the boundary between coding and noncoding RNA
Authors: Suenaga Yusuke
Affiliation: Department of Molecular Carcinogenesis, Chiba Cancer Center Research Institute, 666-2 Nitona, Chuoh-ku, Chiba, Japan
Title: Bottleneck Spatio-Temporal Attention Network for RNA sequences classification of coding or non-coding transcripts
Authors: Yusuke Suenaga
Affiliation: Cancer Genome Center, Chiba Cancer Center Research Institute Chiba