Artificial Intelligence in Oncology
A topical collection in Cancers (ISSN 2072-6694). This collection belongs to the section "Methods and Technologies Development".
Viewed by 315161Editors
2. Institute of Medical Radiology, University Clinic St. Pölten, Karl Landsteiner University of Health Sciences, 3100 St. Pölten, Austria
Interests: metabolic imaging; brain tumors; Warburg effect; reverse Warburg effect; tumor microinvironment; metabolic coupling; energy metabolism; magentic resonance imaging; Förster resonance energy transfer imaging; artificial intelligence; deep learning
Special Issues, Collections and Topics in MDPI journals
Interests: breast MRI; breast tumors
Special Issues, Collections and Topics in MDPI journals
Interests: breast cancer; glioblastoma
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
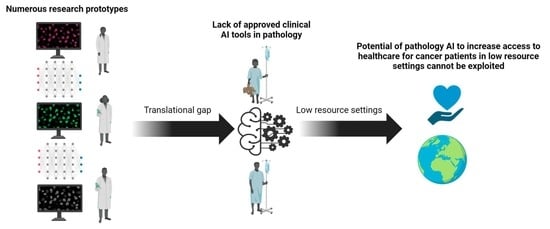
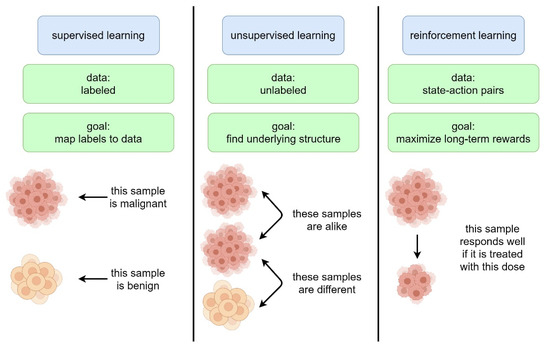
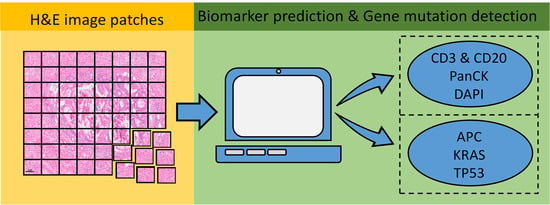
Artificial intelligence (AI) comprises a type of computer science that develops software programs for intelligent execution of tasks or decision making. These approaches allow for bridging the gap between the acquisition of data and its meaningful interpretation. Consequently, artificial intelligence has demonstrated outstanding capabilities for the resolution of a variety of biomedical problems, including cancer, over the past decade.
Machine learning is a subfield of AI and applies mathematical and statistical algorithms to improve computer performance. Deep learning, a subfield of machine learning, is based on artificial neural networks with representation learning that supports automatic feature extraction and provides high flexibility.
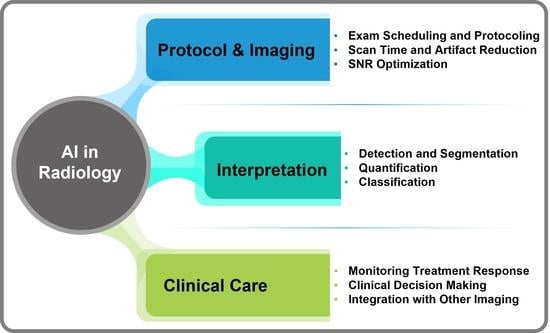
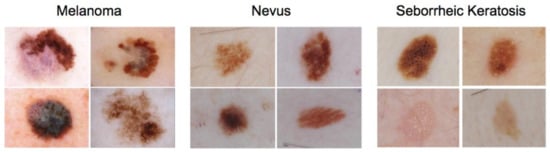
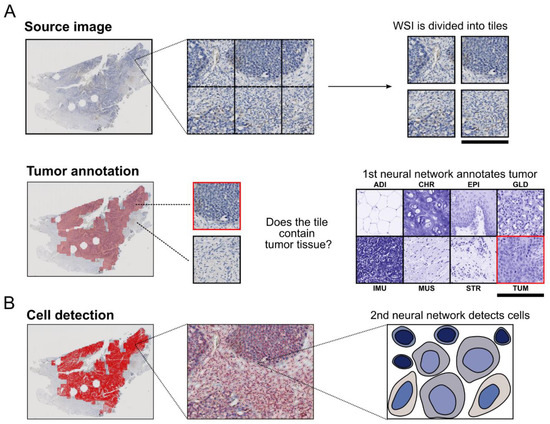
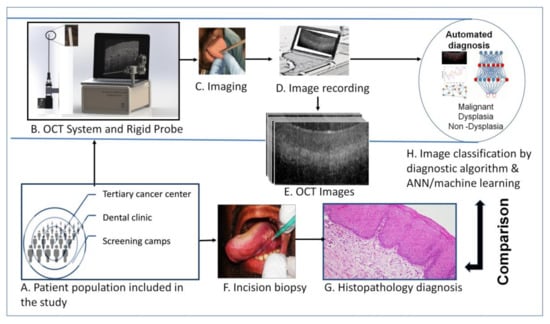
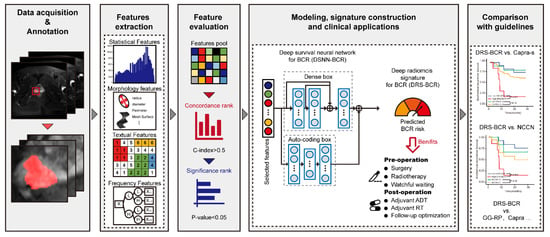
Artificial intelligence can play an essential role in a wide variety of aspects of oncology: tumor detection and segmentation, histopathological diagnosis, tracking tumor development, clinical decision making as well as cancer therapy development and validation or prognosis prediction.
This Topical Collection will highlight several of the above issues. We welcome submissions of research and review articles addressing several facets of AI in both basic and clinical cancer research.
Prof. Dr. Andreas Stadlbauer
Prof. Dr. Anke Meyer-Baese
Dr. Max Zimmermann
Collection Editors
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cancers is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2900 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- artificial intelligence
- deep learning
- machine learning
- oncology
- personalized medicine