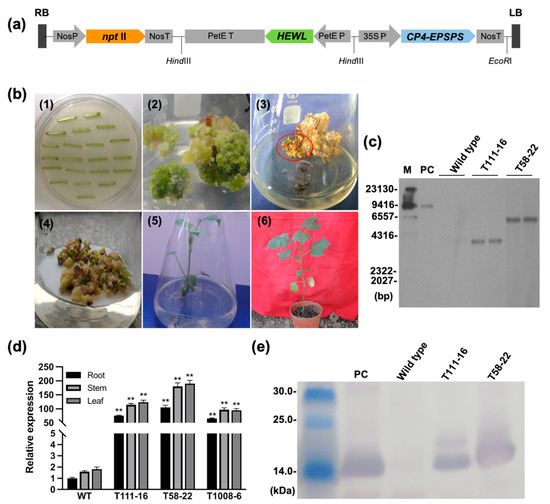
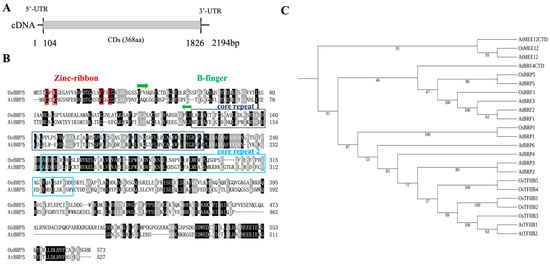
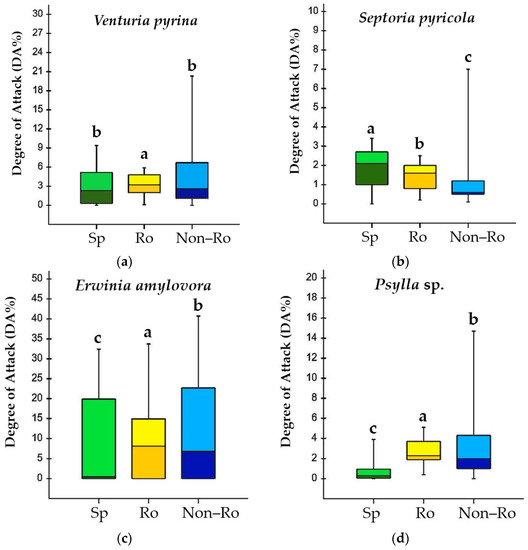
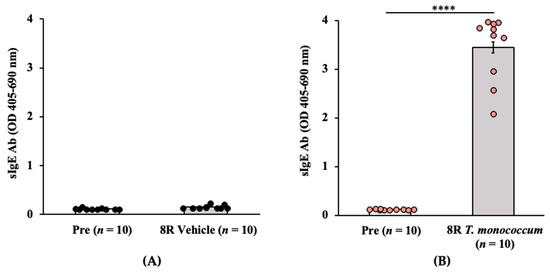
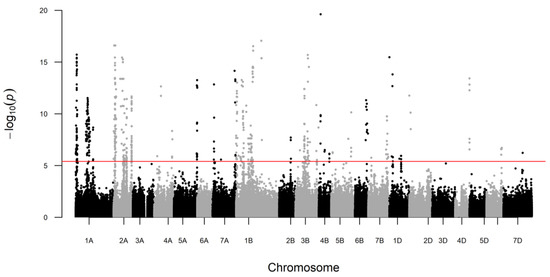
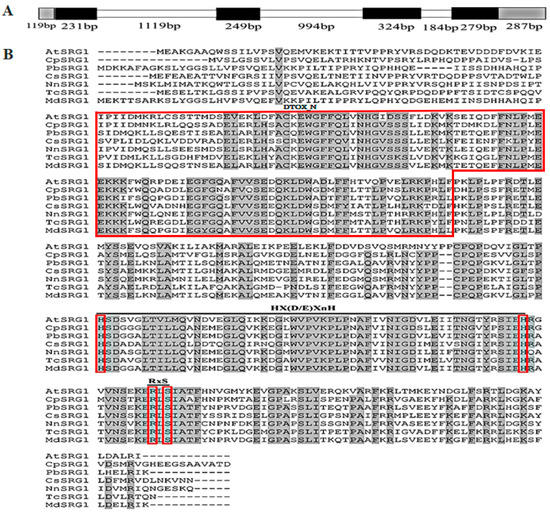
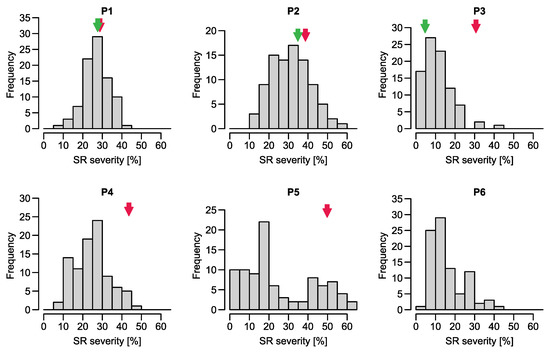
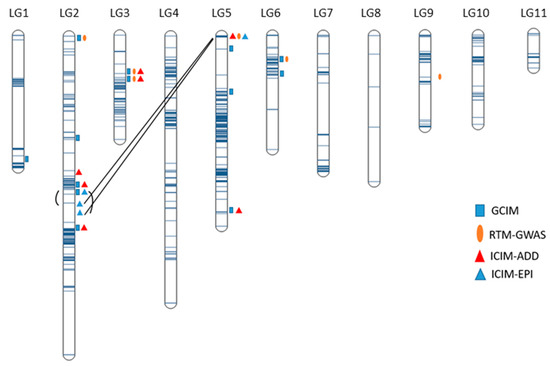
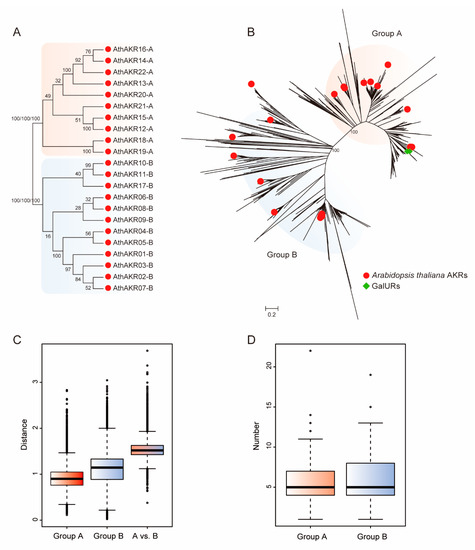
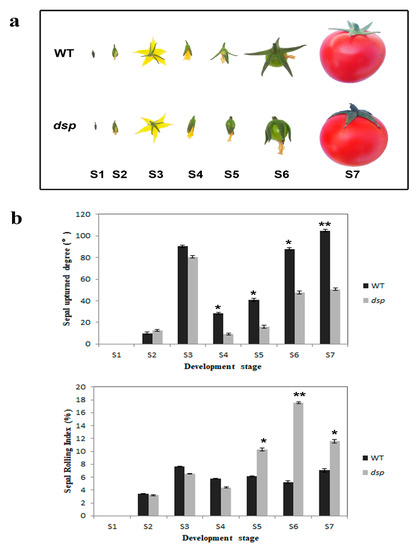
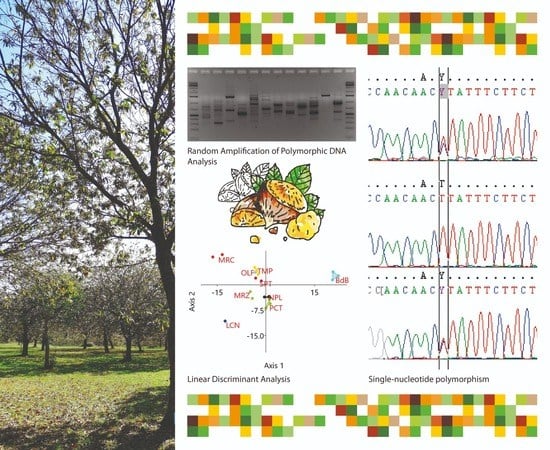
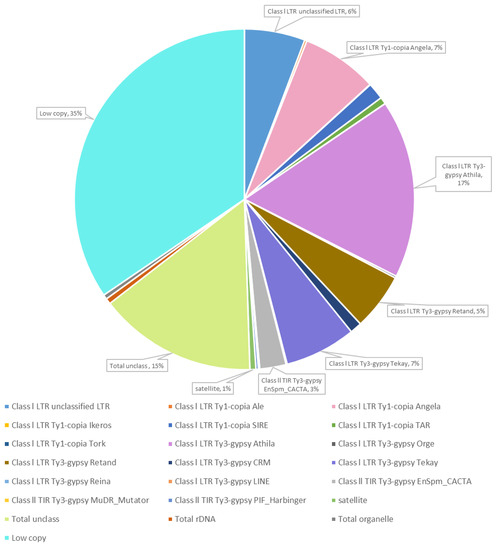
Genetics and Molecular Breeding in Plants
A topical collection in International Journal of Molecular Sciences (ISSN 1422-0067). This collection belongs to the section "Molecular Plant Sciences".
Viewed by 260352Editor
Interests: Prunus breeding; marker-assisted selection; integrating genetic; genomic; transcriptomic; proteomic and epigenetic approaches
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
The development of new plant varieties is a long and tedious process involving the generation of large seedling populations for the selection of the best individuals. While the ability of breeders to generate large populations is almost unlimited, the management, phenotyping (genetic studies), and selection of these seedlings are the main factors limiting the generation of new cultivars. Genomic (DNA) studies for the development of marker-assisted selection (MAS) strategies are particularly useful when the evaluation of the character is expensive, time-consuming, or with long juvenile periods. More recently, proteomic (proteins and enzymes), transcriptomic (RNA), and epigenetic (DNA methylation and histone modifications) studies have been used in the mentioned genomic studies.
Papers submitted to this Topical Collection must report highly novel results and/or plausible and testable new models for the integrative analysis of the different approaches applied to plant breeding, including genetic (phenotyping and transmission of agronomic characters), genomic (DNA regions responsible for the different agronomic characters), proteomic (proteins and enzymes involved in the expression of the characters), transcriptomic (gene expression analysis of the characters), and epigenetic (DNA methylation and histone modifications) approaches for the development of new MAS strategies. In addition, the application of massive sequencing methodologies ("deep-sequencing") of the genome (DNA-Seq) and transcriptome (RNA-Seq), based on lowering the costs of DNA sequencing, could be an additional approach of interest to this Topical Collection.
Dr. Pedro Martínez-Gómez
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. International Journal of Molecular Sciences is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. There is an Article Processing Charge (APC) for publication in this open access journal. For details about the APC please see here. Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- breeding
- genetic
- genomic
- transcriptomic
- proteomic
- epigenetic
- throughput analysis
- assisted selection
Related Special Issue
Planned Papers
The below list represents only planned manuscripts. Some of these manuscripts have not been received by the Editorial Office yet. Papers submitted to MDPI journals are subject to peer-review.
1.
Title: Genome-wide linkage mapping affecting resistance to blank shank in tobacco (Nicotiana tabacum L.)
Author: Lirui Cheng [email protected]
2.
Title: Maize GWAS and its application: Progress and Perspectives
Author: Xiangyuan Wan [email protected]
3.
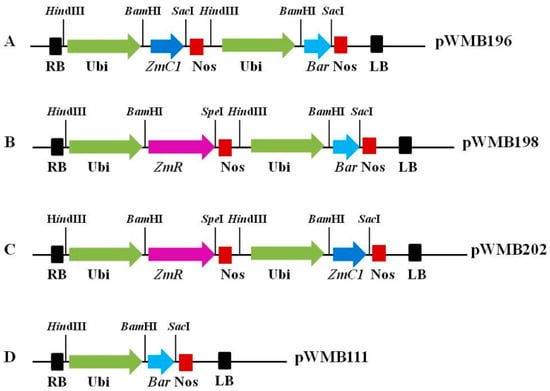
Title: Maize transgenic systems and their molecular breeding application: Progress and Perspectives
Author: Xiangyuan Wan [email protected]
4.
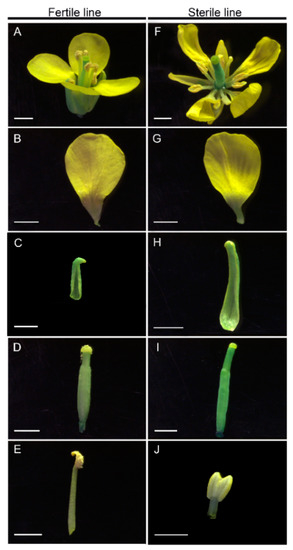
Title: One Maize male sterility gene Ms25
Author: Xiangyuan Wan [email protected]
5.
Topic: R-loci arrangement versus downy and powdery mildew resistance level: a Vitis hybrid survey
Author: Silvia Vezzulli [email protected]